nomogram图,又称列线图,诺模图,可直接用于计算预测的分数
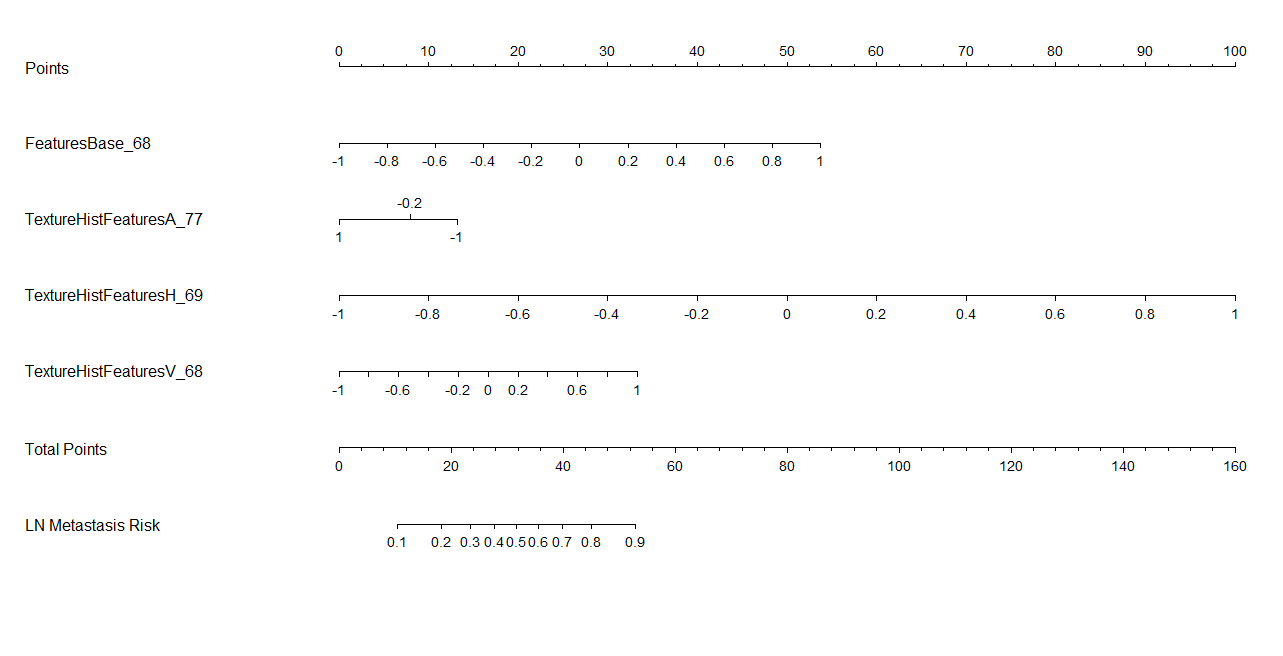 结果如上图所示
参考资料可以去丁香园上找
参考文献:
doi:10.1093/jnci/djv291 Establishment and Validation of Prognostic Nomograms for Endemic Nasopharyngeal Carcinoma
结果如上图所示
参考资料可以去丁香园上找
参考文献:
doi:10.1093/jnci/djv291 Establishment and Validation of Prognostic Nomograms for Endemic Nasopharyngeal Carcinoma
#first need to set path
setwd('E:\Course\cluster') #设置当前工作目录
getwd() #查看当前工作目录
thyroid<- read.table("thyroid.csv",header=FALSE,sep=",") #load
data=thyroid
data=thyroid[,1:10] #太多不拟合
fix(data)
rownames(data)[1]<-"thyroidlabel"
#把1,2的标签换成0 1
for (i in 1:455)
{if(data[i,1]==2)
data[i,1]=0
}
data=data[,-13]
###标签换成yes no的离散的 但是不好用
if(0){
for (i in 1:455)
{if(data[i,13]==0)
data[i,13]='no'
else
data[i,13]='yes'
}}
###
library(rms)
## 第三步 按照nomogram要求“打包”数据,绘制nomogram的关键步骤,??datadist查看详细说明
ddist <- datadist(data)
options(datadist="ddist")
## 第四步 构建模型
## 构建logisitc回归模型
mod <- lrm(data$thyroidlabel~.,data = data)
mod <- glm(data$thyroidlabel ~.,family=binomial(link = "logit"),data = data)
## 绘制logisitc回归的风险预测值的nomogram图
nom <- nomogram(f1, fun= function(x)1/(1+exp(-x)), # or fun=plogis
lp=F, funlabel="LN Metastasis Risk")
plot(nom)
nom <- nomogram(mod,
lp=T,
lp.at = seq(-5,5,by=0.5),
fun=function(x) 1/(1+exp(-x)),
funlabel = 'Risk of metastasis',
fun.at = c(0.05,seq(0.1,0.9,by=0.1),0.95),
conf.int = c(0.1,0.7))
plot(nom,
lplabel = 'Linear Predictor',
fun.side = c(1,1,1,1,3,1,3,1,1,1,1),
label.every = 3,
col.conf = c('blue','green'),
conf.space = c(0.1,0.5),
col.grid = gray(c(0.8,0.95)),
which='shock')
 结果如上图所示
参考资料可以去丁香园上找
参考文献:
doi:10.1093/jnci/djv291 Establishment and Validation of Prognostic Nomograms for Endemic Nasopharyngeal Carcinoma
结果如上图所示
参考资料可以去丁香园上找
参考文献:
doi:10.1093/jnci/djv291 Establishment and Validation of Prognostic Nomograms for Endemic Nasopharyngeal Carcinoma